Abundance boxplot
Usage
abundance_plt(
rec,
taxa_ids = NULL,
type = "boxplot",
transform = "compositional",
scale = 1,
top_n = 20
)
# S4 method for class 'Recipe'
abundance_plt(
rec,
taxa_ids = NULL,
type = "boxplot",
transform = "compositional",
scale = 1,
top_n = 20
)
# S4 method for class 'PrepRecipe'
abundance_plt(
rec,
taxa_ids = NULL,
type = "boxplot",
transform = "compositional",
scale = 1,
top_n = 20
)Arguments
- rec
A Recipe or Recipe step.
- taxa_ids
Character vector with taxa_ids to plot. If taxa_ids is NULL the significant characteristics present in all of the executed methods will be plotted.
- type
Character vector indicating the type of the result. Options: c("boxoplot", "heatmap").
- transform
Transformation to apply. The options include: 'compositional' (ie relative abundance), 'Z', 'log10', 'log10p', 'hellinger', 'identity', 'clr', 'alr', or any method from the vegan::decostand function. If the value is NULL, no normalization is applied and works with the raw counts.
- scale
Scaling constant for the abundance values when transform = "scale".
- top_n
Maximum number of taxa to represent. Default: 20.
Examples
data(test_prep_rec)
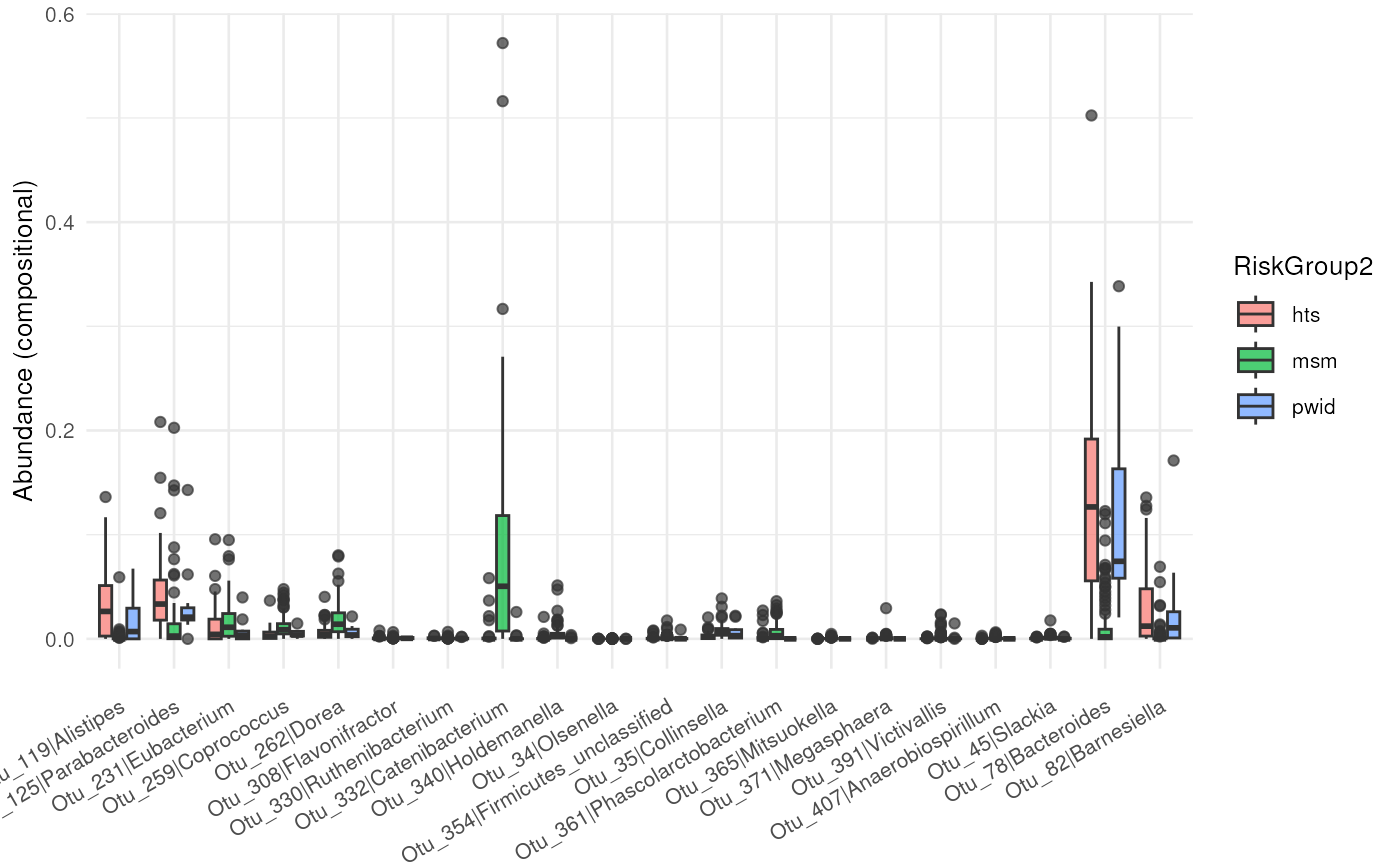
## Running the function returns a boxplot,
abundance_plt(test_prep_rec)
 ## Giving the value "heatmap" to the type parameter, the resulting graph
## a heatmap.
# abundance_plt(test_prep_rec, type = "heatmap")
## By default, those taxa significant in all methods are plotted. If you want
## to graph some determined features, you can pass them as vector through the
## taxa_ids parameter.
# taxa_ids <- c("Otu_96", "Otu_78", "Otu_88", "Otu_35", "Otu_94", "Otu_34")
# abundance_plt(test_prep_rec, taxa_ids = taxa_ids)
# abundance_plt(test_prep_rec, taxa_ids = taxa_ids, type = "heatmap")
## abundance_plt function needs a PrepRecipe. If you pass a a non-prep
## Recipe the output is an error.
data(test_rec)
err <- testthat::expect_error(abundance_plt(test_rec))
err
#> <error/rlang_error>
#> Error in `abundance_plt()`:
#> ! This function needs a PrepRecipe!
#> • Run prep(rec) and then try with abundance_plt()
#> ---
#> Backtrace:
#> ▆
#> 1. ├─testthat::expect_error(abundance_plt(test_rec))
#> 2. │ └─testthat:::expect_condition_matching_(...)
#> 3. │ └─testthat:::quasi_capture(...)
#> 4. │ ├─testthat (local) .capture(...)
#> 5. │ │ └─base::withCallingHandlers(...)
#> 6. │ └─rlang::eval_bare(quo_get_expr(.quo), quo_get_env(.quo))
#> 7. ├─dar::abundance_plt(test_rec)
#> 8. └─dar::abundance_plt(test_rec)
## Giving the value "heatmap" to the type parameter, the resulting graph
## a heatmap.
# abundance_plt(test_prep_rec, type = "heatmap")
## By default, those taxa significant in all methods are plotted. If you want
## to graph some determined features, you can pass them as vector through the
## taxa_ids parameter.
# taxa_ids <- c("Otu_96", "Otu_78", "Otu_88", "Otu_35", "Otu_94", "Otu_34")
# abundance_plt(test_prep_rec, taxa_ids = taxa_ids)
# abundance_plt(test_prep_rec, taxa_ids = taxa_ids, type = "heatmap")
## abundance_plt function needs a PrepRecipe. If you pass a a non-prep
## Recipe the output is an error.
data(test_rec)
err <- testthat::expect_error(abundance_plt(test_rec))
err
#> <error/rlang_error>
#> Error in `abundance_plt()`:
#> ! This function needs a PrepRecipe!
#> • Run prep(rec) and then try with abundance_plt()
#> ---
#> Backtrace:
#> ▆
#> 1. ├─testthat::expect_error(abundance_plt(test_rec))
#> 2. │ └─testthat:::expect_condition_matching_(...)
#> 3. │ └─testthat:::quasi_capture(...)
#> 4. │ ├─testthat (local) .capture(...)
#> 5. │ │ └─base::withCallingHandlers(...)
#> 6. │ └─rlang::eval_bare(quo_get_expr(.quo), quo_get_env(.quo))
#> 7. ├─dar::abundance_plt(test_rec)
#> 8. └─dar::abundance_plt(test_rec)
